- About
- Academics
-
Undergraduate Programs
- Civil and Environmental Engineering
- Architecture and Architectural Engineering
- Mechanical Engineering
- Industrial Engineering
- Energy Resources Engineering
- Nuclear Engineering
- Materials Science and Engineering
- Electrical and Computer Engineering
- Naval Architecture and Ocean Engineering
- Computer Science and Engineering
- Aerospace Engineering
- Chemical and Biological Engineering
-
Graduate Programs
- Civil and Environmental Engineering
- Architecture and Architectural Engineering
- Mechanical Engineering
- Industrial Engineering
- Energy Systems Engineering
- Materials Science and Engineering
- Electrical and Computer Engineering
- Naval Architecture and Ocean Engineering
- Computer Science and Engineering
- Chemical and Biological Engineering
- Aerospace Engineering
- Interdisciplinary Program in Technology, Management, Economics and Policy
- Interdisciplinary Program in Urban Design
- Interdisciplinary Program in Bioengineering
- Interdisciplinary Program in Artificial Intelligence
- Interdisciplinary Program in Intelligent Space and Aerospace Systems
- Chemical Convergence for Energy and Environment Major
- Multiscale Mechanics Design Major
- Hybrid Materials Major
- Double Major Program
- Open Programs
-
Undergraduate Programs
- Research
- Campus Life
- Communication
- Prospective Students
- International Office
Department of Chemical and Biological Engineering, College of Engineering, Seoul National University, Professor Sangwoo Seo's team, Development of a multilevel gene expression regulation system based on RNA-targeting CRISPR-dCas13.
-
Uploaded by
기획협력실
-
Upload Date
2024.07.01
-
Views
1,158
Department of Chemical and Biological Engineering, College of Engineering, Seoul National University, Professor Sangwoo Seo's team,
Development of a multilevel gene expression regulation system based on RNA-targeting CRISPR-dCas13.
- Compared to existing DNA-targeting CRISPR interference systems, gene-specific expression regulation is achievable within the operon.
- Utilization is anticipated for precise regulation of metabolic pathways and maximization of substance production within microorganisms.
- Published online in the prestigious academic journal 'Nature Communications' on June 22nd.

▲(From left) Professor Seo Sangwoo, Department of Chemical and Biological Engineering, Seoul National University, and doctoral student Kim Kiho
The College of Engineering at Seoul National University announced that Professor Sangwoo Seo's research team in the Department of Chemical and Biological Engineering has developed a new synthetic biology methodology capable of precisely controlling gene expression in microbial cell factories.
Previously, tools such as CRISPR-dCas9 or dCpf1 were predominantly used for gene regulation, but they had limitations in simultaneously suppressing the expression of all genes within operon structures where multiple genes are expressed together. This hindered understanding of their effects by independently suppressing expression of each gene within operon structures.

▲In this study, a new tool called CRISPR-dCas13 was utilized for the regulation of gene expression. Genetic information in organisms is typically transcribed from DNA to RNA, and then translated from RNA to proteins, where their functions are exerted. dCas13 targets RNA instead of DNA, unlike traditional gene regulation tools, enabling the suppression of gene expression at the translational level.
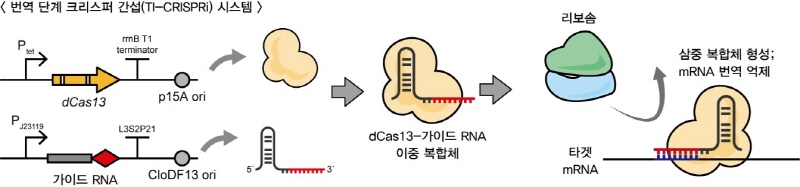
The research team validated that, in regulating the expression of genes expressed in operon structures, they could independently control the expression of each gene within the operon, contrasting with traditional transcriptional inhibition systems. Furthermore, they developed a system capable of predictably adjusting gene translation not only as simple ON/OFF states but across various levels. They constructed an improved guide RNA set by introducing diverse structural changes in the handle region of the guide RNA, enabling suppression of target gene expression from 2.8% to 86.3% with a wide range and uniform distribution.
Using this newly developed translation regulation system, the research team increased the productivity of 3-hydroxypropionic acid, a key raw material for biodegradable plastics, by up to 14 times in E. coli cell factories. Professor Sangwoo Seo stated, "The technology developed in this study, which enables precise control of gene translation, can effectively optimize metabolic processes in microbial cell factories, advancing synthetic biology."

▲This research was recognized for its achievements and published online in the prestigious global scientific journal "Nature Communications" on June 22nd. The study was supported by grants from the Korea Foundation for the Core Technology Development of Synthetic Biology, Next-Generation Bio-Prospective Technology Research Program, Advanced GW Bio-Society Fusion Support Program, Excellent Young Researcher Program, and the Ministry of Oceans and Fisheries' Technology Development Project for Localization of Marine Bio-Industrial Materials.
[Reference]
Title: Tunable translation-level CRISPR interference by dCas13 and engineered gRNA in bacteria
Journal: Nature Communications 15:5319 (2024)
DOI: https://doi.org/10.1038/s41467-024-49642-x
URL: https://www.nature.com/articles/s41467-024-49642-x
[Author]
First Author: Kiho Kim (Doctoral Candidate)
Corresponding author: Professor Sangwoo Seo (Department of Chemical and Biological Engineering, College of Engineering, Seoul National University
Phone: 02-880-2274 Email: swseo@snu.ac.kr