- About
- Academics
-
Undergraduate Programs
- Civil and Environmental Engineering
- Architecture and Architectural Engineering
- Mechanical Engineering
- Industrial Engineering
- Energy Resources Engineering
- Nuclear Engineering
- Materials Science and Engineering
- Electrical and Computer Engineering
- Naval Architecture and Ocean Engineering
- Computer Science and Engineering
- Aerospace Engineering
- Chemical and Biological Engineering
-
Graduate Programs
- Civil and Environmental Engineering
- Architecture and Architectural Engineering
- Mechanical Engineering
- Industrial Engineering
- Energy Systems Engineering
- Materials Science and Engineering
- Electrical and Computer Engineering
- Naval Architecture and Ocean Engineering
- Computer Science and Engineering
- Chemical and Biological Engineering
- Aerospace Engineering
- Interdisciplinary Program in Technology, Management, Economics and Policy
- Interdisciplinary Program in Urban Design
- Interdisciplinary Program in Bioengineering
- Interdisciplinary Program in Artificial Intelligence
- Interdisciplinary Program in Intelligent Space and Aerospace Systems
- Chemical Convergence for Energy and Environment Major
- Multiscale Mechanics Design Major
- Hybrid Materials Major
- Double Major Program
- Open Programs
-
Undergraduate Programs
- Research
- Campus Life
- Communication
- Prospective Students
- International Office
News
Professor Do-Nyun Kim's team at Seoul National University Develops Technology to Predict the Deformation of DNA Origami Structures Induced by DNA-Binding Molecules
-
Uploaded by
대외협력실
-
Upload Date
2024.08.07
-
Views
2,783
Professor Do-Nyun Kim's team at Seoul National University Develops Technology to Predict the Deformation of DNA Origami Structures Induced by DNA-Binding Molecules
Expected to be Utilized in the Design of Mechanochemically Tunable DNA Nanostructures
Necessity of Research Recently, research using DNA origami technology to create nanostructures for advanced bio-convergence fields has been actively conducted. Especially, the technology to utilize environmental changes within the body to modify the shape of structures to perform necessary functions is gaining attention. However, due to the lack of modeling and computer simulation technology that can design these variable mechanisms with understanding, implementing variable technology has relied on repetitive experiments and trial and error. Therefore, there is a need for an effective method to rapidly analyze the deformation mechanisms of structures based on the geometric and mechanical properties of DNA, which change with environmental conditions.
Research Achievements/Expectations Professor Do-Nyun Kim's research team has successfully developed a technology to quickly predict how DNA origami structures will deform depending on the concentration of compounds that bind to DNA. Using this technology, they quantitatively analyzed the geometric and mechanical changes in DNA when combined with a representative DNA-binding molecule, Ethidium Bromide (EtBr), through molecular dynamics simulations. By utilizing this pre-calculated information, they can quickly predict the mechanochemical deformation of various DNA origami structures. This methodology is expected to significantly contribute to the advancement of related technologies by quantifying the changes in DNA properties due to binding with other DNA-binding molecules or various environmental changes, thereby enabling the design of tunable DNA origami structures that can change shape as needed.
Main Text
The College of Engineering at Seoul National University announced that Professor Do-Nyun Kim's research team from the Department of Mechanical Engineering has developed a technology that can quickly predict the mechanochemical shape changes of DNA origami nanostructures.
The results of this study were published on July 31 in the international scientific journal Nature Communications.
DNA origami technology, which uses the self-assembly characteristics of DNA to design and fabricate structures of desired shapes with nanoscale precision, has high applicability and is being actively researched in advanced bio-convergence fields. Especially, technology to utilize environmental changes within the body to alter the shape of structures to perform necessary functions is attracting attention. However, due to the lack of modeling and computer simulation technology that can design these variable mechanisms with understanding, implementing variable technology has relied on repetitive experiments and trial and error. There has been a rising need to study effective methods to rapidly analyze the deformation mechanisms of structures based on the geometric and mechanical properties of DNA, which change with environmental conditions.
Starting from this problem awareness, the research team discovered a way to quickly predict how the shape of DNA origami structures changes depending on the concentration of molecules binding to DNA. The team first quantitatively analyzed the geometric and mechanical changes in DNA caused by binding with a representative DNA-binding molecule, Ethidium Bromide (EtBr), through molecular dynamics simulations. Then, they constructed a relationship between the concentration of binding molecules and changes in DNA properties using this molecular-level calculated information and applied it to DNA structure analysis. Through this process, they were finally able to quickly determine the mechanochemical deformation of DNA origami structures according to changes in EtBr concentration.
This technology can be easily extended to predict DNA property changes due to binding with other DNA-binding molecules. It can also be used to design tunable DNA origami structures that can change shape based on the concentration of binding molecules or according to the type of binding molecules, thereby significantly contributing to the advancement of related DNA nanotechnology and various application research.
Meanwhile, this study, led by Professor Do-nyun Kim of the Department of Mechanical Engineering at Seoul National University and conducted by Research Professor Lee Jae-young and Researcher Kim Yang-kyun, was supported by the Ministry of Science and ICT's Convergence Research and Development Program for Science and Technology Challenges (Artificial Morphogenesis Research Group) and the Mid-career Researcher Support Program.
Results The research team has developed a technology to quickly predict the shape changes of DNA origami structures based on the concentration of molecules binding to DNA. They demonstrated that it is possible to accurately predict the deformation of DNA origami structures when combined with a representative DNA-binding molecule, Ethidium Bromide (EtBr). Furthermore, through comparative analysis with experimental results, they validated the technology's performance in predicting shape and stiffness changes in DNA structures according to EtBr concentration, suggesting broad applicability of this technology.
○ Paper Title: Predicting the effect of binding molecules on the shape and mechanical properties of structured DNA assemblies
○ Authors: Jae Young Lee, Yanggyun Kim, and Do-Nyun Kim
○ Published in: Nature Communications
○ URL: https://www.nature.com/articles/s41467-024-50871-3
Explanation of terms
○ DNA Origami Nanotechnology: A nanotechnology that utilizes the self-assembly properties of DNA to design and fabricate structures of desired shapes with nanoscale precision. This technology involves engineering the base sequences of hundreds of short DNA strands that complement specific parts of a long single DNA strand composed of thousands of bases, thereby achieving the desired shapes and properties.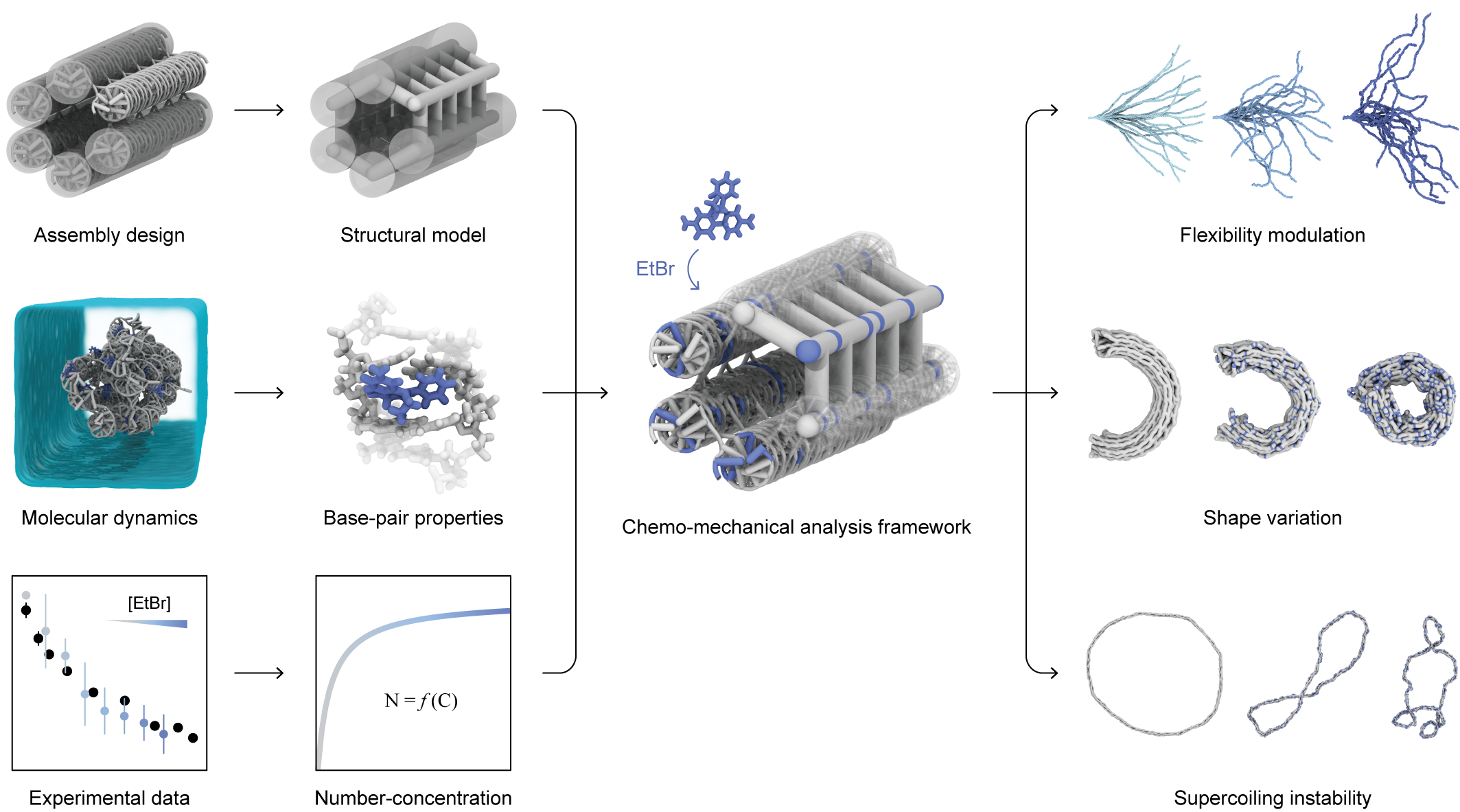
Figure 1. Conceptual Diagram of the Predictive Technology for Mechanochemical Changes in DNA Nanostructures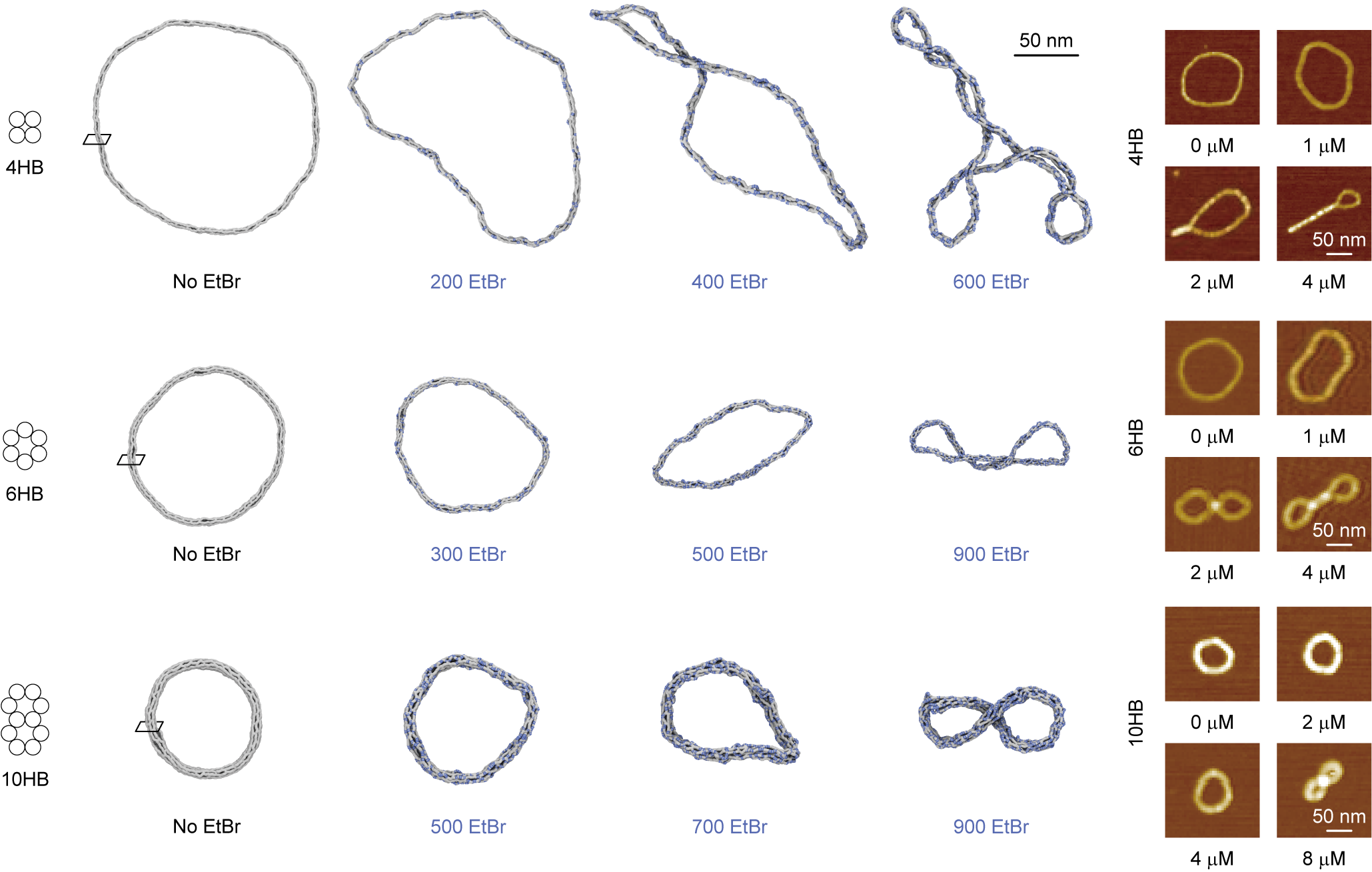
Figure 2. Prediction and Experiment of DNA Ring Structure Supercoiling Phenomenon with Varying EtBr Concentrations
(From left) Professor Do-Nyun Kim, Department of Mechanical Engineering, Seoul National University (corresponding author), Dr. Jae Young Lee (co-first author).
[Contact Information]
Prof. Do-Nyun Kim, Professor, Department of Mechanical Engineering, Seoul National University
Phone: +82-2-880-1647
Email: dnkim@snu.ac.kr
Jae Young Lee, Research Assistant Professor, Institute of Advanced Machinery and Design, Seoul National University
Email: mudge1998@snu.ac.kr